New SMU study may provide insight on how our brains are able to produce so many different types of neurons, which control everything we do
DALLAS (SMU) – SMU (Southern Methodist University) researchers have discovered another layer of complexity in gene expression, which could help explain how we’re able to have so many billions of neurons in our brain.
Neurons are cells inside the brain and nervous system that are responsible for everything we do, think or feel. They use electrical impulses and chemical signals to send information between different areas of the brain, and between the brain and the rest of the nervous system, to tell our body what to do. Humans have approximately 86 billion neurons in the brain that direct us to do things like lift an arm or remember a name.
Yet only a few thousand genes are responsible for creating those neurons.
All cells in the human nervous system have the same genetic information. But ultimately, genes are turned “on” or “off” like a light switch to give neurons specific features and roles. Understanding the mechanism of how a gene is or is not turned on – the process known as gene expression – could help explain how so many neurons are developed in humans and other mammals.
“Studies like this are showing how by unique combinations of specific genes, you can make different specific neurons,” said Adam D. Norris, co-author of the new study and Floyd B. James Assistant Professor in the Department of Biological Sciences at SMU. “So down the road, this could help us explain: No. 1, how did our brain get this complex? And No. 2, how can we imitate nature and make whatever type of neurons we might be interested in following these rules?”
Scientists already have part of the gene expression puzzle figured out, as previous studies have shown that proteins called transcription factors play a key role in helping to turn specific genes on or off by binding to nearby DNA.
It is also known that a process called RNA splicing, which is controlled by RNA binding proteins, can add an additional layer of regulation to that neuron. Once a gene is turned on, different versions of the RNA molecule can be created by RNA splicing.
But before the SMU study was done, which was published in the journal eLife, it was not exactly clear what the logistics of creating that diversity was.
“Before this, scientists had mostly been focused on transcription factors, which is layer No. 1 of gene expression. That’s the layer that usually gets focused on as generating specific neuron types,” Norris said. “We’re adding that second layer and showing that [transcription factors and RNA binding proteins] have to be coordinated properly.
And Norris noted, “this was the first time where coordination of gene expression has been identified in a single neuron.”
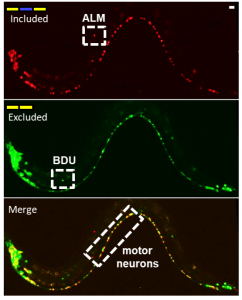
Using a combination of old school and cutting-edge genetics techniques, researchers looked at how the RNA of a gene called sad-1, also found in humans, was spliced in individual neurons of the worm Caenorhabditis elegans. They found that sad-1 was turned on in all neurons, but sad-1 underwent different splicing patterns in different neuron types.
And while transcription factors were not shown to be directly participating in the RNA splicing for the sad-1 gene, they were activating genes that code for RNA binding proteins differently between different types of neurons. It is these RNA binding proteins that control RNA splicing.
“Once that gene was turned on, these factors came in and subtly changed the content of that gene,” Norris said.
As a result, sad-1 was spliced according to neuron-specific patterns.
They also found that the coordinated regulation had different details in different neurons.
“Picture two different neurons wanting to reach the same goal. You can imagine they either go through the exact same path to get there or they take divergent paths. In this study, we’re showing that the answer so far is divergent paths,” said Norris. “Even in a single neuron, there are multiple different layers of gene expression that together make that neuron the unique neuron that it is.”
Norris used worm neurons because “unlike in humans, we know where every worm neuron is and what it should be up to. Therefore, we can very confidently know which genes are responsible for which neural process.
“The very specific details from this study will not apply to humans. But hopefully the principles involved will,” Norris explained. “From the last few decades of work in the worm nervous system, specific genes found to have a specific effect on the worm’s behavior were later shown to be responsible for the same types of things in human nerves.”
The lead author of the study was Morgan Thompson, a graduate student at SMU. Ryan Bixby, Robert Dalton, Alexa Vandenburg — all former or current students in SMU’s Biological Sciences department — also contributed to the study. In addition, John A. Calarco from the University of Toronto, Canada was a co-author.
About SMU
SMU is the nationally ranked global research university in the dynamic city of Dallas. SMU’s alumni, faculty and nearly 12,000 students in seven degree-granting schools demonstrate an entrepreneurial spirit as they lead change in their professions, communities and the world.